Ontology and metadata challenges
Image data is one of the fastest growing outputs of biological research. Owing to the latest advances, the data being produced is larger and more complex making standardisation of image metadata and ontologies is challenging.
Standardisation is critical for effective bioimaging data sharing as well as and cloud-compatible file formats, like the Next Generation File Format, OME-Zarr, as well as toolsets and Application Programming Interfaces (APIs), for interacting and interfacing with the data.
Metadata refers to the information about the data, such as the experimental conditions, sample preparation methods, imaging parameters, etc. Ontologies provide controlled vocabularies for metadata description, facilitating the annotation, sharing, and reuse of imaging data.
Studies annotated with the same metadata schema can be seamlessly integrated with one another, enabling broader comparative analyses and new insights. However, consistent use of same metadata schema is challenging and many different metadata models with varying degrees of compatibility exist.
foundingGIDE work on Ontology and Metadata
Participants in foundingGIDE consortium are major contributors to core biological image metadata models, such as the Open Microscopy Environment (OME) data model and the Recommended Metadata for Biological Images (REMBI) guidelines and major data repositories.
foundingGIDE project partners include research infrastructures representing imaging facilities in their regions, (Euro-BioImaging, National Imaging Facility, Australia, Microscopy Australia, ABiS, Japan with RIKEN the project partner affiliated) data repositories (BioImage Archive, IDR and SSBD:database) and developers of data management systems based on XNAT (AIS XNAT). These repositories, however, are currently not interoperable, leaving the user community to search each in isolation. To support the sharing of image data across these image data repositories, a basic level of compatibility is required.
Deliverables on Ontology and Metadata
We’re excited to share that the project’s first technical deliverables have been reviewed and approved by the European Commission. All the project deliverables are available on Zenodo and freely accessible to the community.
This first set of technical deliverables represents the foundational work to make three global image data repositories (BioImage Archive, IDR and SSBD:database) interoperable by recommending a set of ontologies and a minimal metadata schema to describe the datasets.
Biological Imaging
- D2.1 – Landscape analysis of existing ontologies and recommendations on a set of imaging ontologies
- D6.1 – Report in metadata model overlap and gaps
Preclinical Imaging

foundingGIDE‘s main aim is to enable bioimage data sharing and community efforts around biological and preclinical open image data by enhancing the interoperability across image data resources through reaching consensus agreements on metadata and ontologies used.
Implementing these ontologies and metadata schema in repositories like the BioImage Archive, IDR and SSBD, and creating compatibility with AIS XNAT resources will contribute to making bioimage data FAIR and shareable across the globe, creating a global image data ecosystem of open image data sharing.
Mapping and harmonisation of metadata models between data repositories will allow for functionalities like common search portals and data discoverability mechanisms greatly benefiting for the bioimaging community.
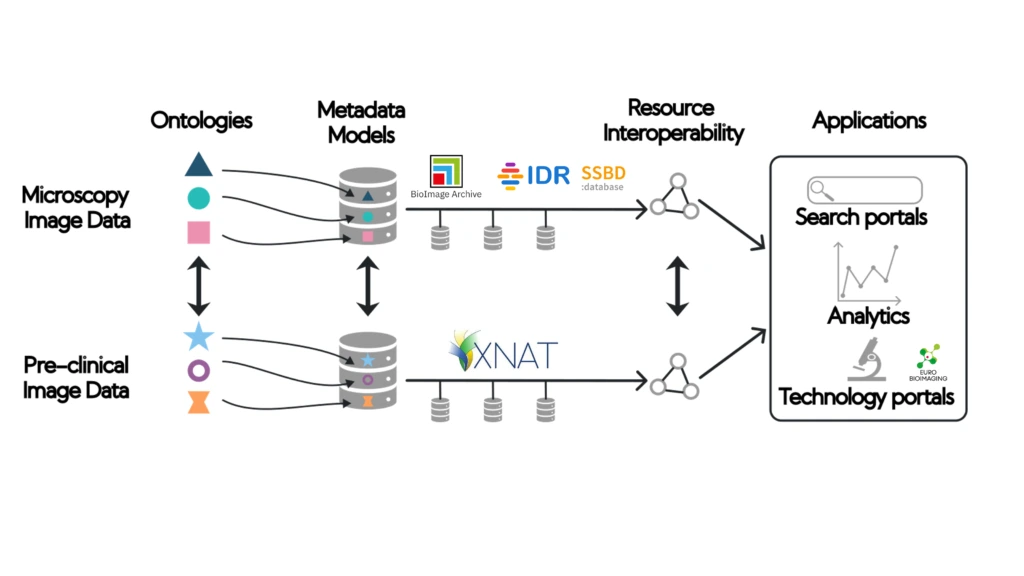
To achieve the main objective of foundingGIDE, a centralised coordination of activities, including community outreach and technical developments, will be required. We will work towards the following aims:
- Establish coordination among global open bioimage data resources
- Increase resource interoperability
- Engage with and provide guidance to community initiatives and end-users
- Plan for sustainability of developed solutions
- Build an ecosystem for new interlinked resources to grow
Find out more about how the bioimage community can get involved in foundingGIDE by visiting our Community page.